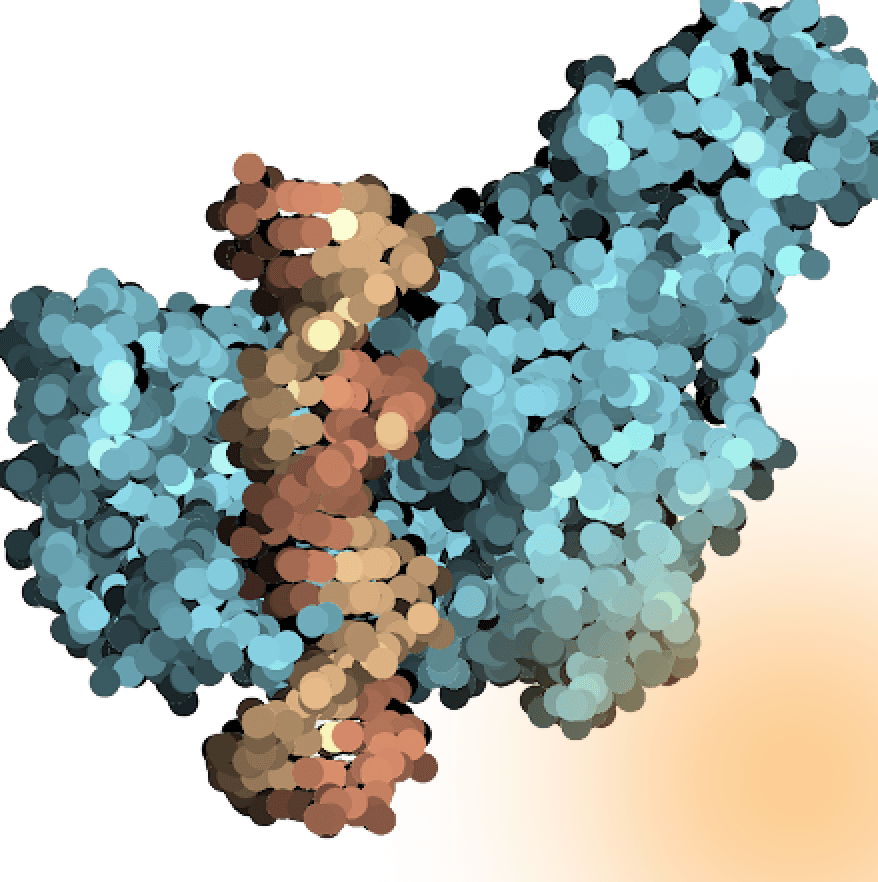
Products
Proprietary engineered enzymes for molecular biology applications

Products
Proprietary engineered enzymes for molecular biology applications

Products
Proprietary engineered enzymes for molecular biology applications
Supreme Phi29
We have engineered multiple phi29 DNA polymerase variants with novel properties that significantly improve the efficiency and versatility of DNA amplification techniques. Identified through Pando’s technology, these mutations offer several advantages over wild-type Phi29 polymerase, including:
We have engineered multiple phi29 DNA polymerase variants with novel properties that significantly improve the efficiency and versatility of DNA amplification techniques. Identified through Pando’s technology, these mutations offer several advantages over wild-type Phi29 polymerase, including:
We have engineered multiple phi29 DNA polymerase variants with novel properties that significantly improve the efficiency and versatility of DNA amplification techniques. Identified through Pando’s technology, these mutations offer several advantages over wild-type Phi29 polymerase, including:
Increased activity
Increased thermostability
Increased manufacturability
Our first product PD-Phi29 is launching in 2025. Contact us for the full data sheet. More products coming soon!
Our first product PD-Phi29 is launching in 2025. Contact us for the full data sheet. More products coming soon!
Increased activity and thermostability
PD-Phi29 is active up to 42°C and creates more DNA faster than the market leader


Figure 1. 100pg of pUC19 was amplified using either a low concentration or high concentration stock of PD-Phi29. The results were compared to the current market best and quantified using a dsDNA dye. The dsDNA production time-course (left graph) was performed at 42°C.
Less bias to GC content variations
PD-Phi29 Whole Genome Amplification (WGA) has less GC-bias than WT or the market best


Figure 2. Whole Genome Amplification was performed on a mix of three bacterial genomes covering a range of GC content, and GC bias of MDA measured via PCR-free Illumina sequencing of the resulting product. Grey bars show the GC distribution of Illumina sequencing data of the input genomic DNA (no WGA), grey MDA performed by wild-type phi29, black using a kit purchased from the market leader, and orange using PD-phi29. The orange line (PD-phi29) is closest to the grey bars, meaning that PD-phi29 has the lowest GC bias. Left panel shows histograms, and the right panel shows the deviation of each WGA sample from the unamplified input DNA. Earth Mover's distance was used to quantify the difference between samples.
Higher success rate in full-plasmid sequencing
PD-Phi29 has a higher success rate for correct and complete full-plasmid sequencing than the market best


Figure 3. Single bacterial colonies were picked, lysed using a thermocycler, and RCA reactions were carried out with the market leading enzyme according to their protocol (grey bar), or with PD-phi29 RCA Kit at the shown temperature, for 1 hour. The amplification products debranched with T7 endonuclease and sequenced using Nanopore. PD-Phi29 has higher yield than the market leading enzyme, and a higher success rate for correct and complete full-plasmid sequencing.
Increased activity and thermostability
PD-Phi29 is active up to 42°C and creates more DNA faster than the market leader


Figure 1. 100pg of pUC19 was amplified using either a low concentration or high concentration stock of PD-Phi29. The results were compared to the current market best and quantified using a dsDNA dye. The dsDNA production time-course (left graph) was performed at 42°C.
Less bias to GC content variations
PD-Phi29 Whole Genome Amplification (WGA) has less GC-bias than WT or the market best


Figure 2. Whole Genome Amplification was performed on a mix of three bacterial genomes covering a range of GC content, and GC bias of MDA measured via PCR-free Illumina sequencing of the resulting product. Grey bars show the GC distribution of Illumina sequencing data of the input genomic DNA (no WGA), grey MDA performed by wild-type phi29, black using a kit purchased from the market leader, and orange using PD-phi29. The orange line (PD-phi29) is closest to the grey bars, meaning that PD-phi29 has the lowest GC bias. Left panel shows histograms, and the right panel shows the deviation of each WGA sample from the unamplified input DNA. Earth Mover's distance was used to quantify the difference between samples.
Higher success rate in full-plasmid sequencing
PD-Phi29 has a higher success rate for correct and complete full-plasmid sequencing than the market best


Figure 3. Single bacterial colonies were picked, lysed using a thermocycler, and RCA reactions were carried out with the market leading enzyme according to their protocol (grey bar), or with PD-phi29 RCA Kit at the shown temperature, for 1 hour. The amplification products debranched with T7 endonuclease and sequenced using Nanopore. PD-Phi29 has higher yield than the market leading enzyme, and a higher success rate for correct and complete full-plasmid sequencing.
Increased activity and thermostability
PD-Phi29 is active up to 42°C and creates more DNA faster than the market leader


Figure 1. 100pg of pUC19 was amplified using either a low concentration or high concentration stock of PD-Phi29. The results were compared to the current market best and quantified using a dsDNA dye. The dsDNA production time-course (upper graph) was performed at 42°C.
Less bias to GC content variations
PD-Phi29 Whole Genome Amplification (WGA) has less GC-bias than WT or the market best


Figure 2. Whole Genome Amplification was performed on a mix of three bacterial genomes covering a range of GC content, and GC bias of MDA measured via PCR-free Illumina sequencing of the resulting product. Grey bars show the GC distribution of Illumina sequencing data of the input genomic DNA (no WGA), grey MDA performed by wild-type phi29, black using a kit purchased from the market leader, and orange using PD-phi29. The orange line (PD-phi29) is closest to the grey bars, meaning that PD-phi29 has the lowest GC bias. Left panel shows histograms, and the right panel shows the deviation of each WGA sample from the unamplified input DNA. Earth Mover's distance was used to quantify the difference between samples.
Higher success rate in full-plasmid sequencing
PD-Phi29 has a higher success rate for correct and complete full-plasmid sequencing than the market best


Figure 3. Single bacterial colonies were picked, lysed using a thermocycler, and RCA reactions were carried out with the market leading enzyme according to their protocol (black bar), or with PD-phi29 RCA Kit at the shown temperature, for 1 hour. The amplification products debranched with T7 endonuclease and sequenced using Nanopore. PD-Phi29 has higher yield than the market leading enzyme, and a higher success rate for correct and complete full-plasmid sequencing.
Interested in distributing Pando products?
Interested in distributing Pando products?
Interested in distributing Pando products?
Interested in getting a free sample?
Interested in getting a free sample?
Interested in getting a free sample?

Ready to Find or Enhance Your Enzymes?

What We Offer
Contact Us
134 Coolidge Avenue, Watertown, Massachusetts 02472, United States
COPYRIGHT © 2025 PANDO BIOSCIENCE - ALL RIGHTS RESERVED.

What We Offer
Contact Us
134 Coolidge Avenue, Watertown, Massachusetts 02472, United States
COPYRIGHT © 2025 PANDO BIOSCIENCE - ALL RIGHTS RESERVED.

What We Offer
Contact Us
134 Coolidge Avenue, Watertown, Massachusetts 02472, United States
COPYRIGHT © 2025 PANDO BIOSCIENCE - ALL RIGHTS RESERVED.